Research
- Structural Chemistry (Experimental and Theoretical)
- Photomolecular Science
- Molecular Spectroscopy
- Biophysical Chemistry
Research Overview
From a molecule to a living cell: Structural chemistry bridges material and life sciences
Whenever we look into the mechanism of how this material world operates, we come across with the question of structure and dynamics of molecules and molecular systems. For example, if we want to understand the process of a chemical reaction at the molecular level, we need to know the structure and dynamics of reaction intermediates that exist only in time-scales of picosecond (10-12 s) or even femtosecond (10-15 s). Our research interests extend over the following four major fields of physical chemistry, photomolecular science, structural chemistry, molecular spectroscopy and biophysical chemistry (Fig. 1). Under strong interactions among researches in these fields, we are aiming at developing new methodology for studying the structure and dynamics of molecules and molecular systems, and eventually, discovering and elucidating new molecular phenomena that will lead to a new concept of basic scientific importance.
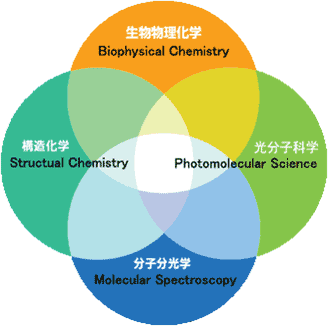
Fig. 1. Research Area
Dynamic structure of reaction intermediates (structural chemistry)
In the course of picosecond time-resolved Raman spectroscopic studies of S1 trans-stilbene, a prototype photochemical reaction intermediate, we noticed that the central C=C stretch Raman band was much broader than the same band in the ground state. We have developed a band-shape theory based on the Kubo-Anderson formalism, in order to clarify the possible correlation between the fast vibrational dephasing (broad Raman band) and high reactivity of the reaction intermediate. We treat this problem under the asymmetric limit of the two frequency exchange scheme, in which S1 trans-stilbene occasionally polarizes into a zwitterion by solvent fluctuation and comes back very fast (Fig. 2, solvent induced dynamic polarization). The analysis based on this theory shows that, in hexane, W1=2.7×1012 s-1 and S1 trans-stilbene polarizes into the zwitterion in every 370 fs on an average. The same analysis in different alkanes and at different temperature has revealed that the polarization rate W1 has a good linear relationship with the overall rate of photoisomerization. From these results, we propose a new model of photoisomerization reaction that proceeds via dynamic polarization of S1 trans-stilbene into a zwitterion.
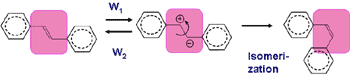
Fig. 2. Solvent-induced dynamic polarization of S1 trans-stilbene in solution
We also applied this theory to thermal reactions in ground electronic states. For instance, we have shown that tert-butyl-alcohol in 1 M sulphuric acid dissociates into the carbocation in 5 ps on an average and come back in 400 fs.
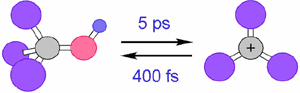
Fig. 3. Formation/annihilation Dynamics of the carbocation in 1 M sulphuric acid
Ultrafast photoinduced stepwise double proton-transfer reaction of 2-aminopyridine/acetic acid complex (photomolecular science)
Photoinduced double proton transfer (PDPT) reaction is of considerable interest as an elementary process of the so called gproton relayh, which plays crucial roles in a variety of biological processes. There has been a lot of discussion on how the two protons move, concerted or stepwise, in PDPT. We investigated the PDPT reaction of 2-aminopyridine/acetic acid complex by picosecond time-resolved fluorescence spectroscopy. We have proved that the reaction proceeds in a stepwise fashion; immediately after photoexcitation, the proton of acetic acid moves to the ring nitrogen of 2-aminopyridine to form an ion-pair intermediate and then, with a 5 ps time constant in hexane, the amino proton moves to acetic acid to complete PDPT. We have also found that the second proton transfer is thermally activated but that there is no H/D isotope effect on the activation energy. This fact indicates that the motion of proton is not involved in the reaction coordinate, which is not well accounted for by any existing theory. We are now considering the effect of the solvent induced dynamic polarization of the ion-pair intermediate on the rate of the second proton transfer reaction.
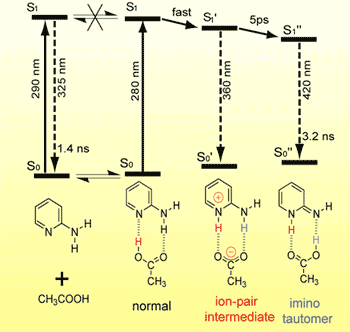
Fig. 4. Ultrafast photoinduced double proton transfer reaction of 2-aminopyridine/acetic acid complex
Development of Infrared Electroabsorption (IREA) Spectroscopy and Its Application to Structural Studies (molecular spectroscopy)
Infrared electroabsorption (IREA) spectroscopy provides unique higher-order vibrational information on molecular species existing in liquids and solutions. Species having distinct dipole moments are detected distinctly by IREA spectra and their dipole moments are determined separately. On account of technical difficulties, however, IREA was so far applicable only to wavenumber regions higher than 3000 cm-1. We developed a new IREA system based on a dispersive spectrometer and the AC-coupling detection scheme. It is capable of detecting infrared absorbance changes as small as 10-8 in ΔA and enables the measurements of IREA in the fingerprint region. Fig. 2 shows the IREA spectra of a 1,4-dioxane solution of N-methylacetamide observed with varying angle χthe angle between the incident infrared electric vector and the direction of the applied external electric field. The high S/N ratio in these spectra enabled us to analyze the χ dependence by SVD (Singular Value Decomposition) and determine the dipole moments of the dimer as well as the monomer of N-methylacetamide in 1,4-dioxane. The analysis shows that the dimmer has a head-to-tail structure. We were thus able to determine the structure of an aggregated species in solution, which is prohibitively difficult with other spectroscopic/diffraction methods.
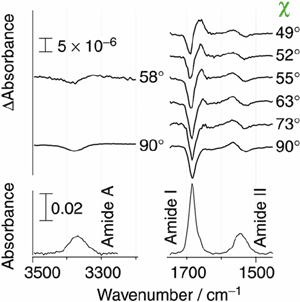
Fig. 5. Infrared electroabsorption spectrum of N-methylacetamide in 1,4-dioxane
Discovery of the gRaman Spectroscopic Signature of Lifeh and in vivo Raman tracing of a spontaneous cell death process of a yeast cell (biophysical chemistry)
Con-focal Raman microspectroscopy provides a powerful nondestructive means with high time, space and molecular specificity, which are very essential for in vivo molecular-level studies of living cells. We recently discovered a Raman band that sharply reflects the metabolic activity of mitochondria in a living S. Pombe cell and called it "the Raman spectroscopic signature of life". By using this signature, we are able to quantitatively trace the process of cell growth and death at the cellular level. We also found that the Raman spectroscopic signature of life disappears concomitantly with a sudden appearance of a particle called "dancing body" in a vacuole of a budding yeast (S. cerevisiae) cell and that the appearance of the dancing body necessarily results in a loss of the vacuole and, eventually, in an inevitable cell death. Molecular-level changes in organelles accompanying this spontaneous cell death process, which we suspect is a programmed cell death, are traced successfully by time-resolved Raman mapping (Figure 5). We are now able to study the life and death of single living cells at the molecular level using the well-established approach of physical chemistry.
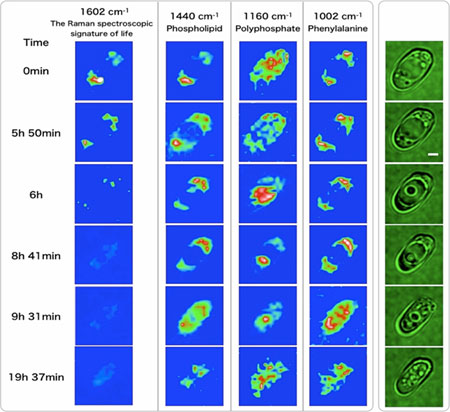
Fig. 6. Time-resolved Raman images (blue) and microscopic images (green) of a living/dying S. cerevisiae cell